and the distribution of digital products.
How DeepSee Helps Us See Deeper into the Oceans than Ever Before
:::info Authors:
(1) Adam Coscia, Georgia Institute of Technology, Atlanta, Georgia, USA ([email protected]);
(2) Haley M. Sapers, Division of Geological and Planetary Sciences, California Institute of Technology, Pasadena, California, USA ([email protected]);
(3) Noah Deutsch, Harvard University Cambridge, Massachusetts, USA ([email protected]);
(4) Malika Khurana, The New York Times Company, New York, New York, USA ([email protected]);
(5) John S. Magyar, Division of Geological and Planetary Sciences, California Institute of Technology Pasadena, California, USA ([email protected]);
(6) Sergio A. Parra, Division of Geological and Planetary Sciences, California Institute of Technology Pasadena, California, USA ([email protected]);
(7) Daniel R. Utter, [email protected] Division of Geological and Planetary Sciences, California Institute of Technology Pasadena, California, USA ([email protected]);
(8) John S. Magyar, Division of Geological and Planetary Sciences, California Institute of Technology Pasadena, California, USA ([email protected]);
(9) David W. Caress, Monterey Bay Aquarium Research Institute, Moss Landing, California, USA ([email protected]);
(10) Eric J. Martin Jennifer B. Paduan Monterey Bay Aquarium Research Institute, Moss Landing, California, USA ([email protected]);
(11) Jennifer B. Paduan, Monterey Bay Aquarium Research Institute, Moss Landing, California, USA ([email protected]);
(12) Maggie Hendrie, ArtCenter College of Design, Pasadena, California, USA ([email protected]);
(13) Santiago Lombeyda, California Institute of Technology, Pasadena, California, USA ([email protected]);
(14) Hillary Mushkin, California Institute of Technology, Pasadena, California, USA ([email protected]);
(15) Alex Endert, Georgia Institute of Technology, Atlanta, Georgia, USA ([email protected]);
(16) Scott Davidoff, Jet Propulsion Laboratory, California Institute of Technology, Pasadena, California, USA ([email protected]);
(17) Victoria J. Orphan, Division of Geological and Planetary Sciences, California Institute of Technology, Pasadena, California, USA ([email protected]).
:::
Table of Links4 Studying Deep Ocean Ecosystem and 4.1 Deep Ocean Research Goals
4.3 Design Challenges and User Tasks
- 5.1 Map View
- 5.2 Core View
5.3 Interpolation View and 5.4 Implementation
6 Usage Scenarios and 6.1 Scenario: Pre-Cruise Planning
- 6.2 Scenario: On-the-Fly Decision-Making
7 Evaluation and 7.1 Cruise Deployment
8 Conclusions and Future Work, Acknowledgments, and References
ABSTRACTScientists studying deep ocean microbial ecosystems use limited numbers of sediment samples collected from the seafloor to characterize important life-sustaining biogeochemical cycles in the environment. Yet conducting fieldwork to sample these extreme remote environments is both expensive and time consuming, requiring tools that enable scientists to explore the sampling history of field sites and predict where taking new samples is likely to maximize scientific return. We conducted a collaborative, user-centered design study with a team of scientific researchers to develop DeepSee, an interactive data workspace that visualizes 2D and 3D interpolations of biogeochemical and microbial processes in context together with sediment sampling history overlaid on 2D seafloor maps. Based on a field deployment and qualitative interviews, we found that DeepSee increased the scientific return from limited sample sizes, catalyzed new research workflows, reduced long-term costs of sharing data, and supported teamwork and communication between team members with diverse research goals.
CCS CONCEPTS• Human-centered computing → Scientific visualization; Visual analytics; Geographic visualization; Visualization systems and tools; • Applied computing → Earth and atmospheric sciences.
KEYWORDSData visualization, scientific visualization, visual analytics, design study, deep ocean research.
\ ACM Reference Format:
\ Adam Coscia, Haley M. Sapers, Noah Deutsch, Malika Khurana, John S. Magyar, Sergio A. Parra, Daniel R. Utter, Rebecca L. Wipfler, David W. Caress, Eric J. Martin, Jennifer B. Paduan, Maggie Hendrie, Santiago Lombeyda, Hillary Mushkin, Alex Endert, Scott Davidoff, and Victoria J. Orphan. 2024. DeepSee: Multidimensional Visualizations of Seabed Ecosystems. In Proceedings of the CHI Conference on Human Factors in Computing Systems (CHI ’24), May 11–16, 2024, Honolulu, HI, USA. ACM, New York, NY, USA, 16 pages. https://doi.org/10.1145/3613904.3642001
1 INTRODUCTIONExploration and research on deep-sea benthic ecosystems, ranging from the expansive abyssal plains to the physico-chemically heterogeneous extreme environments of hydrothermal vents and methane seeps, has significantly advanced in the past two decades. These advances arise from massive amounts of data produced by the development of new technologies for autonomous seafloor mapping and imaging, physico-chemical sensing, biological measurements, and sample archival [31]. Through multidisciplinary teams of scientists providing complementary data sets on the geology, geochemistry, macro- and microbiology of these remote ecosystems, deep ocean researchers can now access 2D topographic maps and color photomosaics of the seafloor, allowing for the relation of point-source seafloor sample collections (e.g. sediment cores, rock, animal, and water samples) with their appropriate environmental context at centimeter to kilometer spatial scales.
\ However, while 2D maps of spatial locations of samples are valuable, the field currently lacks visualization tools which extend into the 3rd dimension, i.e. within the subseafloor. This presents several fundamental data visualization challenges around exploring associated physico-chemical and biological data of samples within their spatial context. The samples are sediment cores – pointwise geospatial and temporal (i.e., multidimensional) data consisting of hundreds of microbial species and physio-chemical parameters – that are spread sparsely across several hundred meters of seafloor. Researchers perform fieldwork to collect these sediment cores from the deep ocean during expeditions, an expensive and time-consuming task that requires extensive planning beforehand and usually results in a very limited number of samples. Seeing prior sampling history to decide where to sample next can help maximize time spent conducting expensive fieldwork. Further, it is unclear how scientist’s existing research workflows might change by introducing new visualization capabilities, both in planning before fieldwork as well as in tactical decision making in the field as new samples are collected. Our goals are to develop a system that solves the visualization challenges in exploring spatial trends between sediment cores in the context of their environment, deploy the system to measure the impacts on scientists’ workflows conducting deep ocean fieldwork, and reflect on broader design guidance for visualizing multidimensional sample data collected during fieldwork expeditions.
\ We present a collaborative design study [43] conducted with a team of scientific researchers studying deep ocean ecosystems (Sect. 3). We first characterized their research goals, preexisting workflows and data practices, leading us to synthesize design challenges and user tasks (Sect. 4) for a system that can help scientists visualize spatial trends between cores in context of the environment, integrate core and map data at multiple size and time scales in a single interface, and increase the scientific return (long-term usability and value of the data) from the limited number of samples available by predicting unseen values in unsampled locations.
\ From our task abstraction, we developed DeepSee (Fig. 1, Sect. 5), an open-source[1], interactive workspace for scientists to upload sediment core data and map images and see their sampling history displayed across multiple connected views simultaneously. Interactive maps of the seafloor between centimeter and kilometer resolution (Fig. 1A) are labeled with information about previous dives as well as collected samples. Alongside these maps, DeepSee displays 2D visualizations that show parameter gradients as a function of depth and interactive 3D visualizations of data interpolations in the space between samples (Fig. 1B). The data interpolations can be run in real time, allowing scientists to “see” below the seafloor and determine the most likely places to collect high-value samples. To support decision making, DeepSee provides annotation tools on the maps for taking notes, useful for communicating findings and planning future dives. Finally, DeepSee is portable and requires no internet access, empowering scientists to use DeepSee on field expeditions in remote environments.
\ To assess the impact of our approach on solving a real-world problem, we evaluated DeepSee with the same expert collaborators involved in the design process (Sect. 7). First, we conducted an initial field deployment of DeepSee aboard a research cruise to test its capabilities and illustrate which user tasks were accomplished in the field. Then, we interviewed each team member and collected feedback on the deployment around several areas of deep ocean fieldwork research that DeepSee impacted. We learned that:
\ (1) Fluid interaction [16] and aggregation of data visualized at multiple scales can help scientists increase the scientific return of limited samples by enabling new ways of asking questions about the data;
\ (2) Integrating 2D and 3D data in the same interface can catalyze new research workflows by surfacing new potential targets for future sampling and reducing the long-term cost of data preparation; and
\ (3) Modular visualizations can help team members with different roles and diverse research goals to solve specific tasks as they decide where and which samples to collect.
\ From these findings, we synthesized principles for designing future visualization systems in other fieldwork-driven domains:
\ (1) Prioritize data integration as a user task when designing, helping users track data being added on the fly in the field;
\ (2) Visualize physical data in context of the environment, fostering stronger understanding and communication of insights;
\ (3) Combine data types to bridge micro and macro scale analysis, increasing scientific return on limited samples; and
\ (4) Design interactive visualizations to aid mental modeling, helping scientists understand and explore complex processes
\ Overall, the combination of fluid interaction, tightly integrated data, and modular visualizations provided deep insight into the data that helped researchers build a stronger intuition about the spatial
\
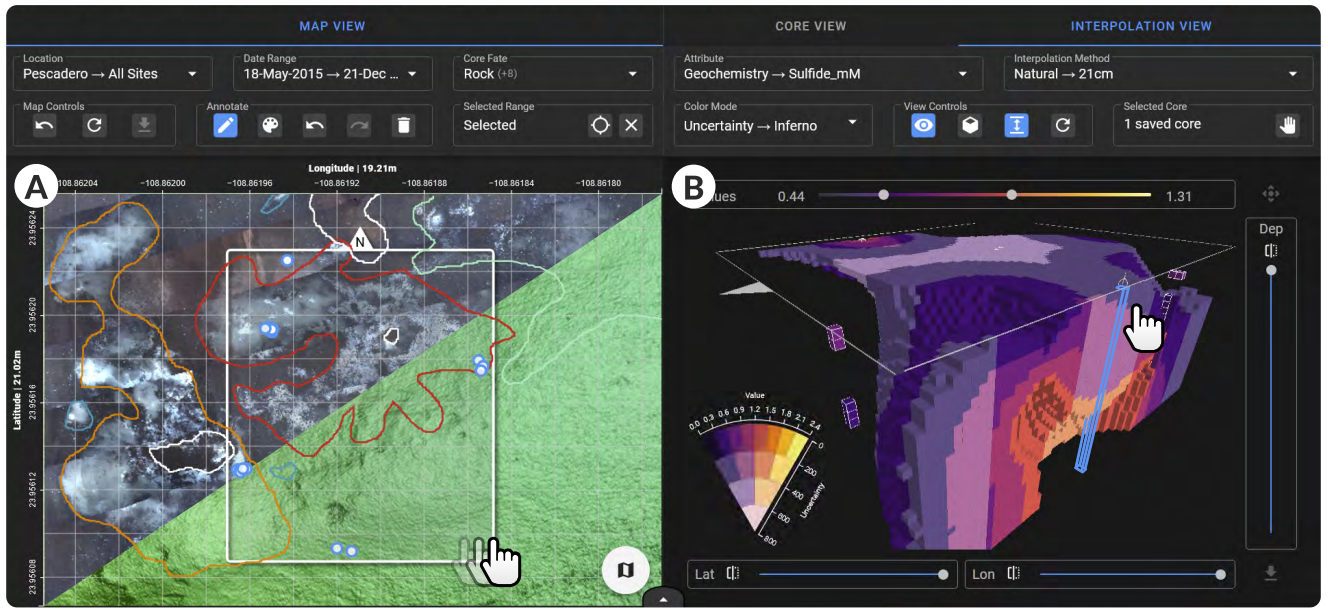
\ distributions of environmental sample data, ultimately facilitating more informed future sample selection.
\ In summary, the contributions of this paper are: (1) a characterization of the scientific process and design considerations for visualizing deep ocean ecosystems following a user-centered design process; (2) DeepSee, an interactive workspace that visualizes the history of deep sea sediment sampling for selecting future sample site locations; and (3) reflections based on interviews with scientists that deployed DeepSee discussing the lessons learned and broader impacts of leveraging data visualizations to support fieldwork-driven scientific research.
\
:::info This paper is available on arxiv under CC BY 4.0 DEED license.
:::
[1] DeepSee code: https://github.com/orphanlab/DeepSee
- Home
- About Us
- Write For Us / Submit Content
- Advertising And Affiliates
- Feeds And Syndication
- Contact Us
- Login
- Privacy
All Rights Reserved. Copyright , Central Coast Communications, Inc.